Search for dense subgraphs
Search for dense subgraphs (or corrupted cliques) is a very common problem arising in bioinformatics (e.g., co-expression of genes) and studies of social interactions (e.g., recommendation services).
We decided to make an algorithm for dense subgraphs search available as an independent tool. One can download IgRepertoireConstructor archive and run DSF for search of dense subgraphs. Details of DSF usage can be found in manual.
Input / output
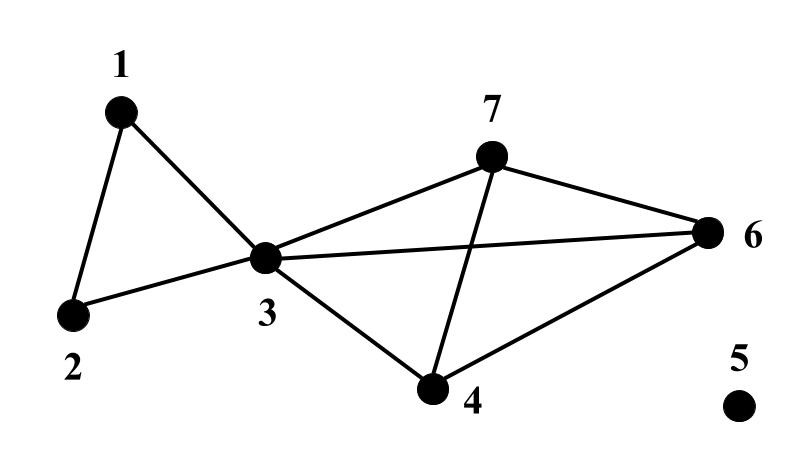
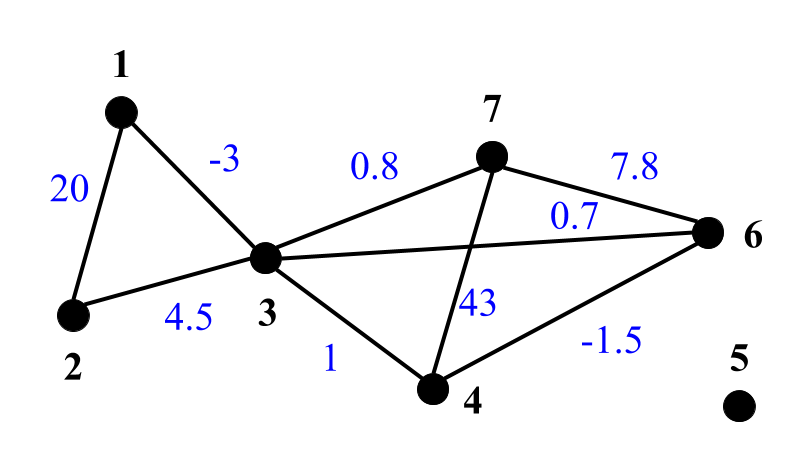
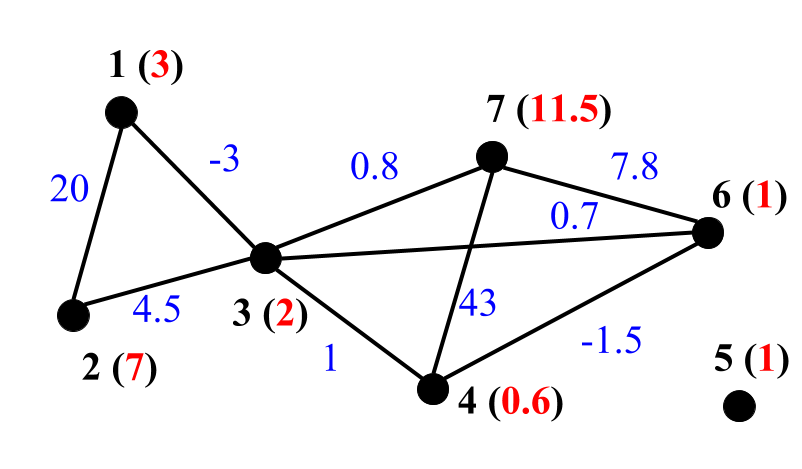
DSF takes indirected graph in GRAPH format as an input. Below are examples representation of unweigthed (left), edge weighted (middle) and edge & vertex weighted (right) graphs in GRAPH format:
- Unweighted: the first line shows number of vertices (7) and number of edges (9). Each of the next lines contains vertex neighbours. E.g., the second line notes that vertex 1 is connected with vertices 2 and 3.
- Edge weighted: the first line shows number of vertices (7), number of edges (9) and description 001 that denotes edge weighted graph. Each of the next lines contains vertex neighbours and weights of corresponding edges (highlighted in blue) for graph vertices. E.g., the second line notes that vertex 1 is connected with vertex 2 by edge of weigth 20 and vertex 3 by the edge of weigth -3.
- Edge & vertex weighted: the first line shows number of vertices (7), number of edges (9) and description 011 that denotes edge & vertex weighted graph. Each of the next lines contains vertex weight (highlighed in red), its neighbours and weights of corresponding edges (highlighted in blue). E.g., the second line notes that vertex 1 of weight 3 is connected with vertex 2 by edge of weigth 20 and vertex 3 by the edge of weigth -3.
Output of DSF is a decomposition of an input graph where the identical ids are assigned for vertices from the same dense subgraph.
Example of DSF work
Density of constructed subgraph can be tuned via parameter -f (minimum edge fill-in in dense subgraphs).
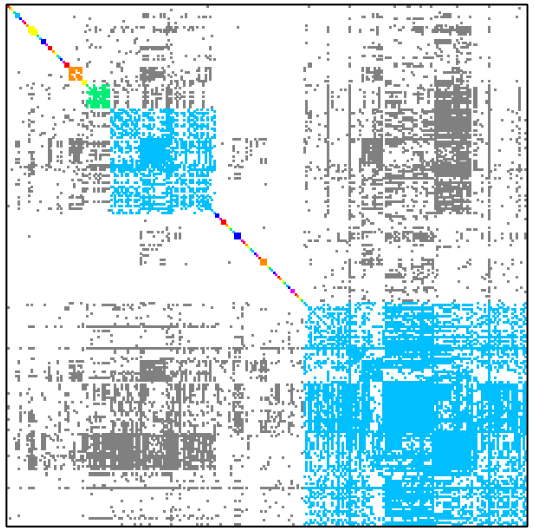
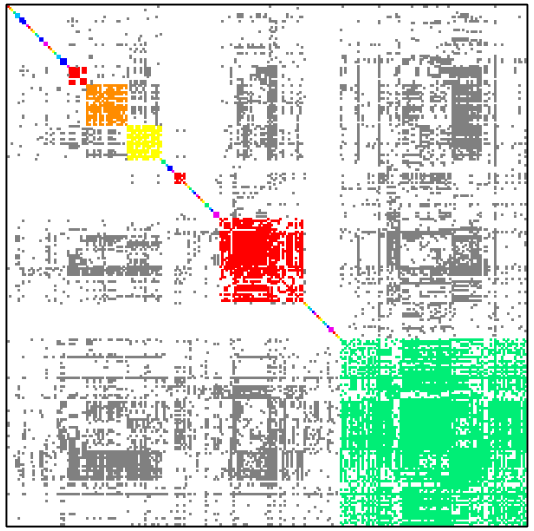
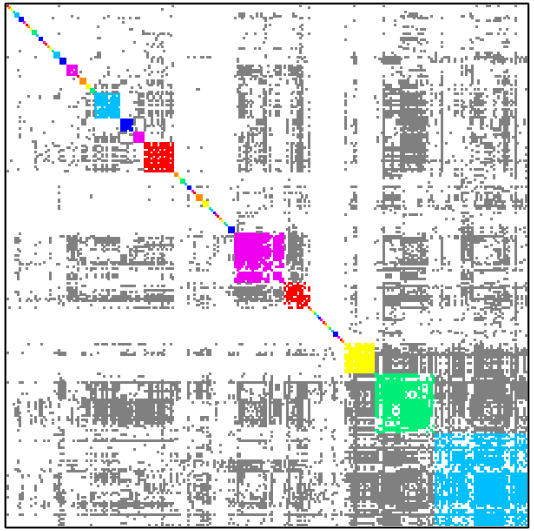
Table below shows how dense subgraph decomposition varies depending of minimal edge fill-in value.
|
|
| Minimal edge fill-in = 0.3 | Minimal edge fill-in = 0.4 |
| # dense subgraphs = 55 # trivial subgraphs = 40 size of maximal subgraph = 157 | # dense subgraphs = 61 # trivial subgraphs = 46 size of maximal subgraph = 104 |
|
|
| Minimal edge fill-in = 0.5 | Minimal edge fill-in = 0.7 |
| # dense subgraphs = 60 # trivial subgraphs = 43 size of maximal subgraph = 87 | # dense subgraphs = 68 # trivial subgraphs = 42 size of maximal subgraph = 43 |
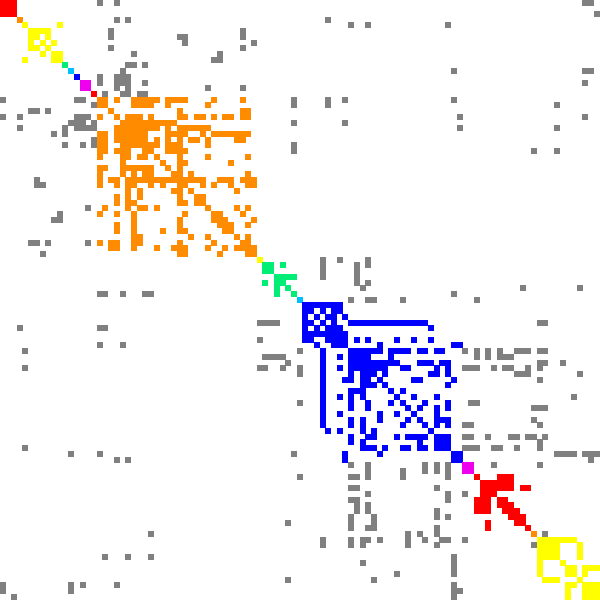
Here is an example of DSF work for finding recommendations. Graph polbooks shows books about USA politics that were bought by the same customer on Amazon. Dense subgraphs in such graph present books that can be recommended together.
|
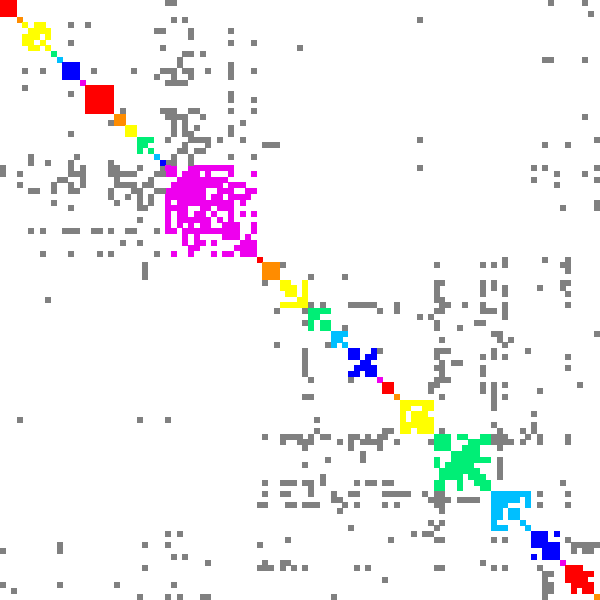
|
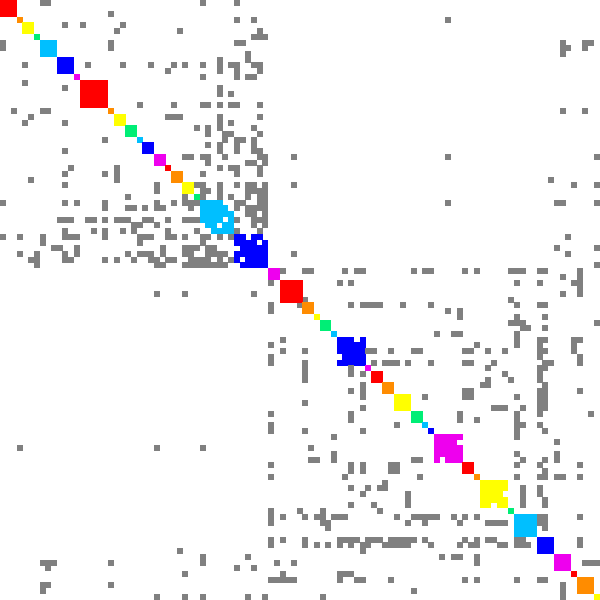
|
| Minimal edge fill-in = 0.2 | Minimal edge fill-in = 0.5 | Minimal edge fill-in = 0.7 |
| # dense subgraphs = 17 # trivial subgraphs = 8 size of maximal subgraph = 28 | # dense subgraphs = 30 # trivial subgraphs = 11 size of maximal subgraph = 16 | # dense subgraphs = 45 # trivial subgraphs = 16 size of maximal subgraph = 6 |
Citation and feedback
Your comments, suggestions and bug reports are very welcome. If you use DSF in your research, please cite our paper:
- Yana Safonova, Stefano Bonissone, Eugene Kurpilyansky, Ekaterina Starostina, Alla Lapidus, Jeremy Stinson, Laura DePalatis, Wendy Sandoval, Jennie Lill, and Pavel A. Pevzner. IgRepertoireConstructor: a novel algorithm for antibody repertoire construction and immunoproteogenomics analysis. Bioinformatics. 2015 Jun 15; 31(12): i53-i61. doi: 10.1093/bioinformatics/btv238.